Development of a data-driven multi-omics module repertoire for data analysis and interpretation
Background
Advances in technology have facilitated the generation of deep systems-level immune profiling data. Efforts to characterize these datasets rely on the availability of well-annotated sources of prior knowledge, such as gene modules for enrichment analysis. Modules consist of groups of co-expressed analytes and provide a stable framework for identifying and annotating signatures to understand broader patterns of immune responses. While certain components of immune responses can be explained through single assay profiling, the complex interplay between various molecular factors is dependent upon multiple assays, motivating the need for multi-omics modules.
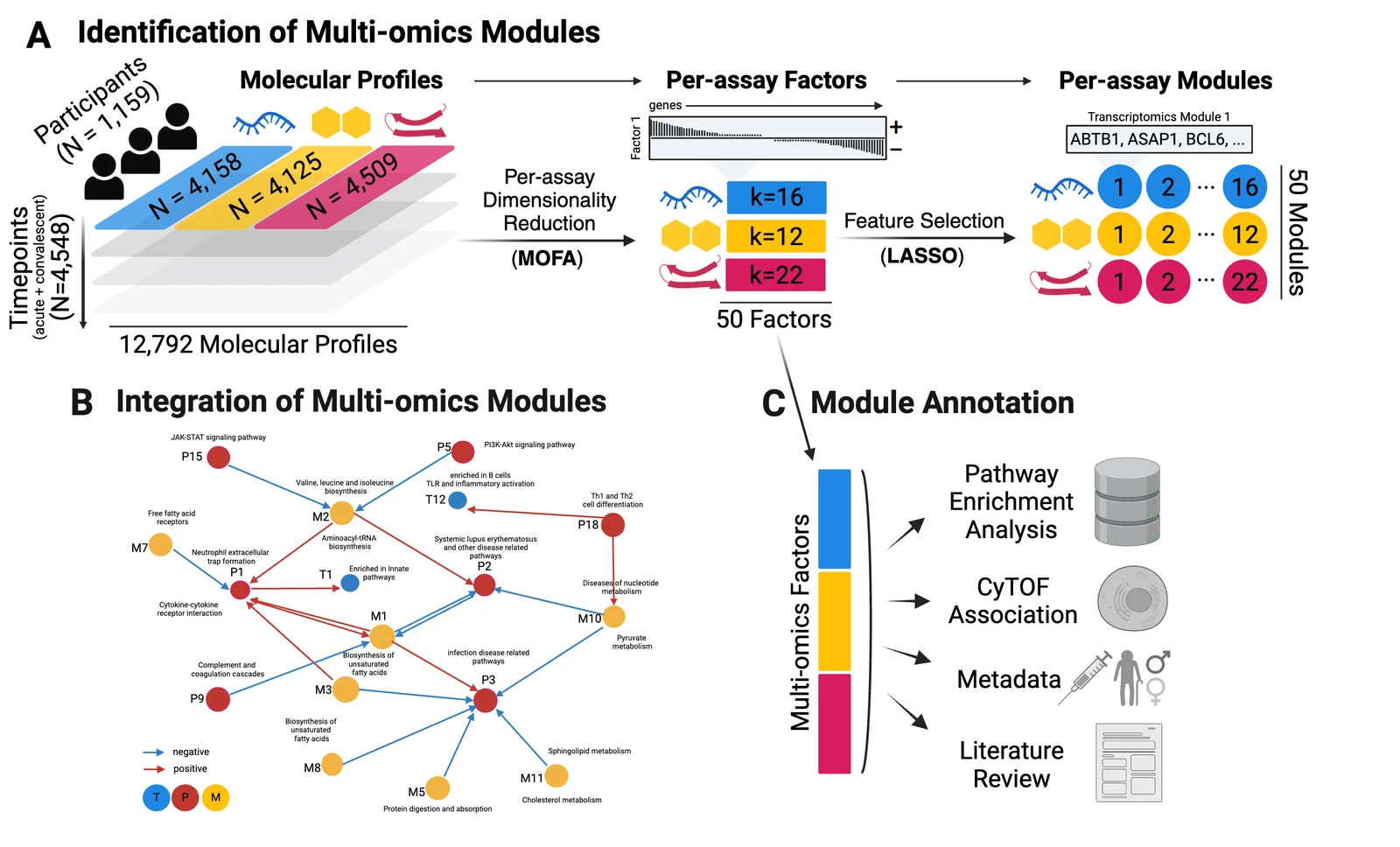
Figure 1: Module construction overview. A) Multi-omics profiles from 1,159 participants over 4,548 timepoints are integrated via per-assay dimensionality reduction into 50 multi-omics factors. Factors are subsequently converted into modules via LASSO feature selection. B) Multi-omics modules are integrated via inter-omic network. C) Multi-omics modules are annotated via enrichment analysis and association of the corresponding multi-omics factors. Modules are characterized via pathway enrichment analysis, CyTOF and metadata association, and manual literature review.
Results
As part of the Human Immunology Project Consortium (HIPC), we have developed Multi-Omics Modules version 1 (MOMod1), a set of data-driven multi-omics modules designed to serve as a stable and reusable framework for downstream enrichment analysis of multi-omics data. MOMod1 encapsulates 12,792 paired transcriptomic, proteomic, and metabolomic molecular profiles from acute and convalescent periods for 1,159 hospitalized SARS-CoV-2 participants, represented as 50 diverse immune programs. We present a thorough investigation into the underlying biological processes represented in each module. We evaluated cross-omics predictive relationships and found strong concordance between proteomic and metabolomic modules, whereas predictions involving transcriptomic modules were weaker, underscoring the complexity of multi-omics integration and the need for advanced modeling strategies. Finally, we validate MOMod1 with multiple external datasets spanning both viral and bacterial infections.
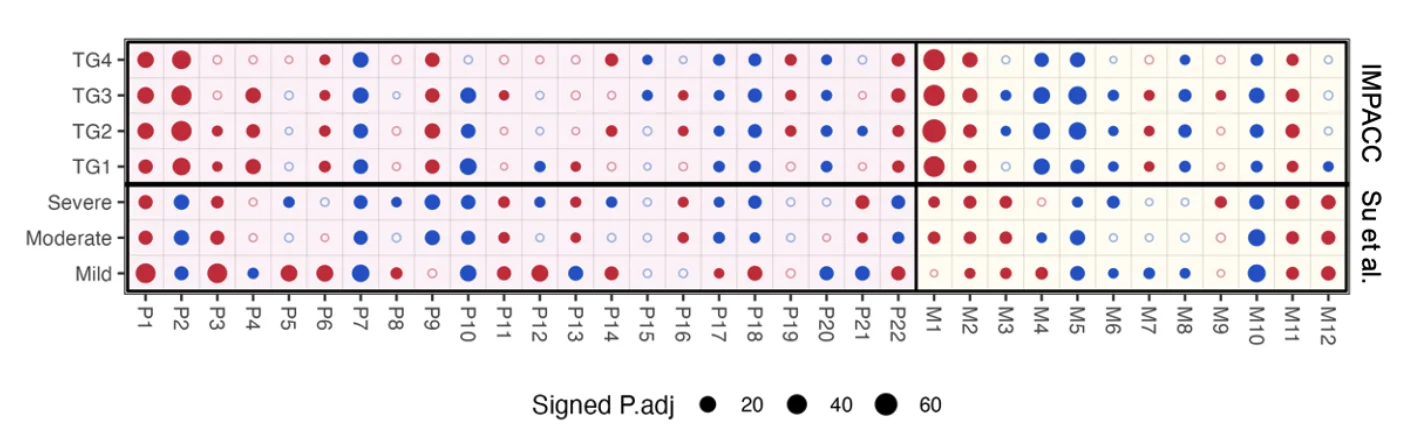
Figure 2: Validation of MOMod1 on COVID-19 Multi-omics Datasets. Comparison of proteomic and metabolomic profiles taken from the IMPACC study (top) and Su et al.’s study (bottom). All points show the corresponding signed adjusted log p-values for comparisons of geometric means of module scores. IMPACC comparisons compare immune states from within disease course trajectory groups (least severe - TG1 < TG2 < TG3 < TG4 – most severe), comparing the earliest acute hospitalized measurement (case) to the latest convalescent measurement (control). Su et al.’s comparisons compare individuals marked as having Mild, Moderate, or Severe COVID-19 (case) to healthy individuals (control). Multi-omics modules are split by assay, with proteomics modules P1-P22 on the left and metabolomics modules (M1-M12) on the right.
Resource: MOMod1 (Version 1.0, Last Updated: March 04, 2025)
MOMod1 can be utilized with immune profiling data consisting of transcriptomic, metabolomic, and proteomic profiles. The following is a list of analytes included in each module.
- List of all MoMod1 modules: MoMod1_list_20250304.txt
- Metabolite ID mapping information: metabolite_id_mapping.txt
Please note: Module definitions and included analytes are subject to revision as the resource evolves. To track changes over time, refer to the version number and update date.
Feel free to contact Pramod Shinde (pshinde@lji.org) and Jian Xing (jian.xing@yale.edu) with any queries.